
Duplex Stabilization
Duplex Stabilization
Duplex stabilization refers to the chemical modification of nucleic acid bases to improve thermal stability, binding strength, and nuclease resistance. Modified bases such as 2-Amino-dA, pdC, pdU and Locked Nucleic Acids (LNA) improve oligonucleotide performance through unique structural changes. The improved heat resistance makes this modification particularly useful for a variety of molecular biology applications, including antisense oligonucleotides (ASOs), siRNA, and PCR probes.
Modification
Chemical Change
∆Tm* / per mod
Benefits
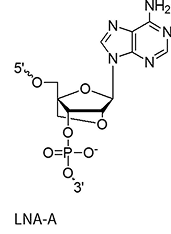
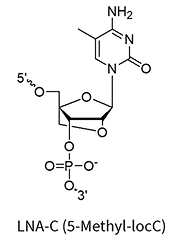
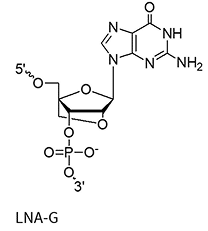
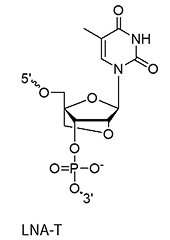
LNA bases (+A/+mC/+G/+T)
A methylene bridge connecting the 2′-O and 4′-C atoms, locking the ribose in a C3′-endo conformation
+2-10°C
This structural rigidity significantly increases duplex stability, improves mismatch discrimination, and enhances nuclease resistance. It also allows for shorter oligo designs and is fairly cheap.
* Values may vary depending on your oligo design
Modification
Chemical Change
∆Tm* / per mod
Benefits
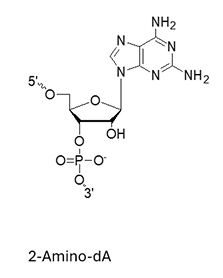
2-Amino-dA
Forms an additional hydrogen bond with thymine, increasing the number of hydrogen bonds from two to three
~+1°C
Strengthens duplex stability and enhances base-pairing specificity. This extra bonding also helps prevent A-G wobble mismatches, By reinforcing the correct A-T pairing, 2-Amino-dA reduces the likelihood of such errors.
* Values may vary depending on your oligo design
Modification
Chemical Change
∆Tm* / per mod
Benefits
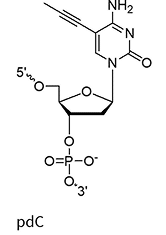
pdC
(propynyl-deoxycytidine)
Propynyl replaces the methyl group on cytosine (pdC) C-5 position
~+2.8°C
The C-5 propynyl modification improves duplex stability by increasing the melting temperature. The propynyl group, which is planar and more hydrophobic than the methyl it replaces, strengthens base stacking and reduces water accessibility, further stabilizing the duplex.
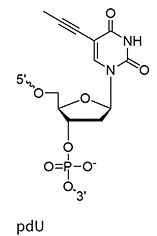
pdU
(propynyl-deoxyuridine)
Propynyl replaces the methyl group on uracil (pdU) C-5 position
~+1.7°C
Same as above
* Values may vary depending on your oligo design
Modification
Chemical Change
∆Tm* / per mod
Benefits
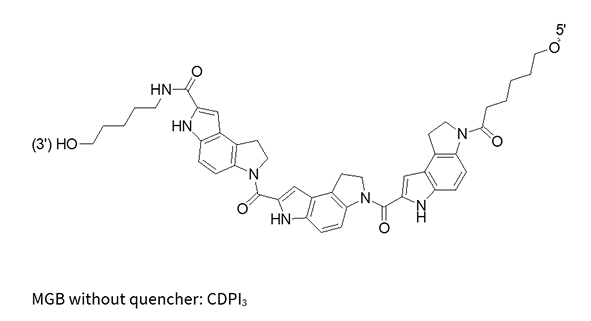
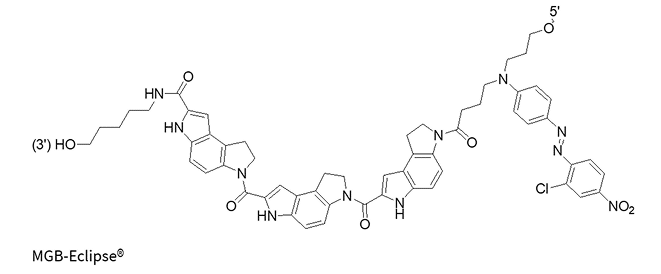
CDPI3
(MGB without quencher)
MGB-Eclipse®
MGBs, such as CDPI3, are crescent-shaped molecules that bind non-covalently to the DNA minor groove. Their shape allows them to precisely conform to the curve of the DNA, stabilizing the interaction through van der Waals forces and hydrophobic effects.
Significantly increases the Tm of DNA duplexes
The MGB molecule forms highly stable complexes with target DNA, allowing for shorter probe designs for better specificity and mismatch discrimination, thus improving quenching efficiency. MGB probes are particularly effective at stabilizing A/T-rich duplexes compared to G/C-rich duplexes.
* Values may vary depending on your oligo design
We are hiring
Are you ready to combine science with creativity? At Ella Biotech, you’ll join a close-knit team dedicated to advancing the life sciences. You’ll find a vibrant, respectful workplace where teamwork drives innovation and fun is always part of the mix.
Bring your talents to our team where every day is an exciting opportunity to make a difference in biotechnology and advance discovery. Let’s create the building blocks of tomorrow – together!

Contact us
„*“ zeigt erforderliche Felder an
